Data Slicer
The Data Slicer provides an interface which allows users to get subsections of either VCF (VCFtools) or BAM (SAMtools) files based on genomic coordinates.
In the Ensembl GRCh37 browser, you can access the Data Slicer from the tools link in the menu bar at the top of every page.
The input interface is shown below:
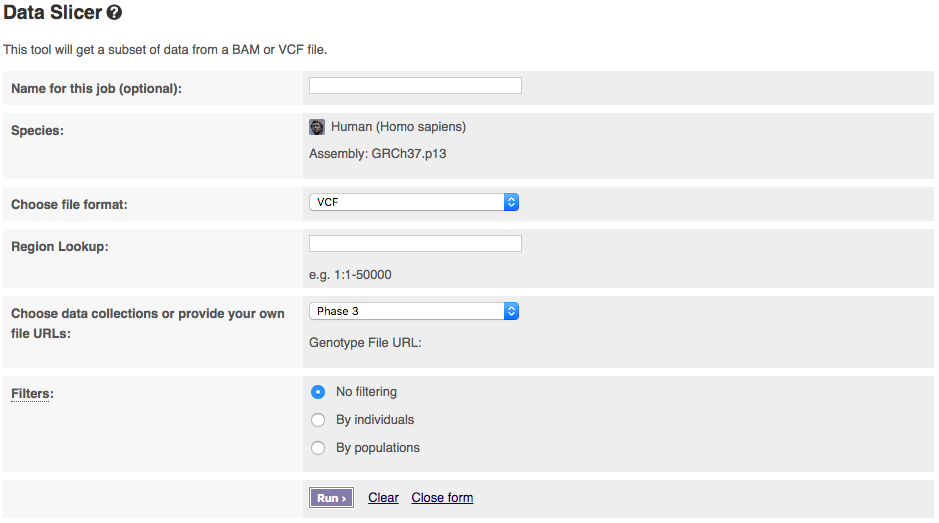
The tool allows you to pick which phase of the 1000 Genomes Project you want to get data from. If you have a publicly visible VCF file and corresponding tabix index (.tbi) in the same folder, you could get data from these by selecting “Provide file URLs”.
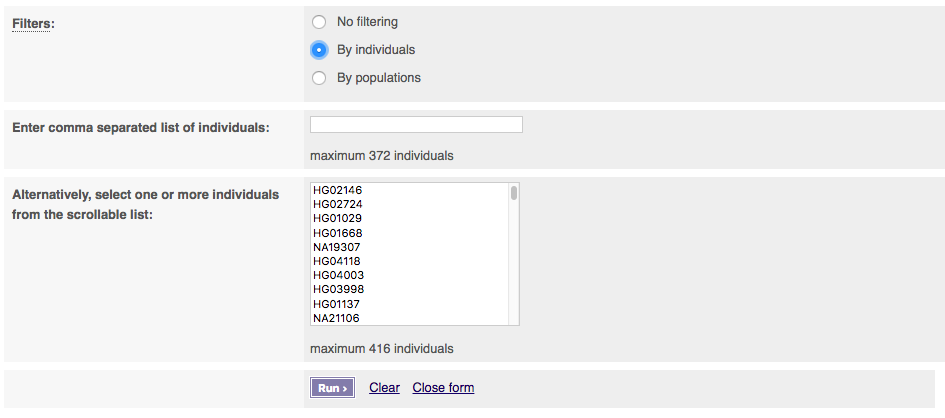
You can select filtering by either individuals or populations. Select one to get extra options. To select multiple populations or individuals please hold the ctrl key (on windows/linux) or the cmd key (macs).
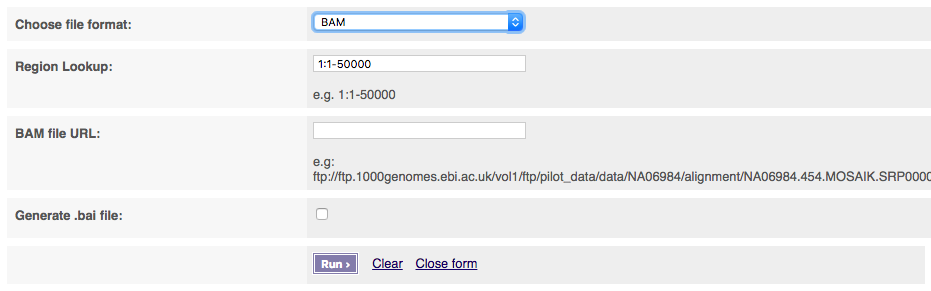
If you select BAM files, you must provide a publicly visible URL for your BAM file which must be accompanied by BAM index (.bai) of the same name. Please note that this service will only work for other BAM files over http.
After clicking “Run” the system produces your final file.
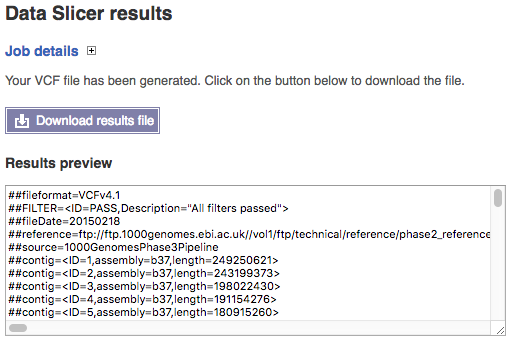
This presents a link to download your file slice which should contain both the location you requested and the name of the original in its name. You also get a summary of the first few lines of the file in the window so you can check it has produced something close to expectation.
There is also documentation for our Variation Pattern finder which looks at shared patterns of variation between individuals from VCF files.
If you have any questions about this system please email info@1000genomes.org