FORGE2
Please note that, since the conclusion of the 1000 Genomes Project, FORGE has been updated to FORGE2, which is hosted at the Altius Institute.
FORGE
This is a set of Forge results generated by analysis of GWAS SNP sets obtained from the GWAS catalog (Downloaded 07-08-2013). The examples are divided roughly into categories of phenotypes that have some broad similarity in terms of aetiology. Click on a thumbnail to be taken to the full PDF of the result. There is also an online gallery of the alternative interactive chart output available at http://iandunham.github.io/gallery/.
Jump to disease section:
- Autoimmune Diseases
- Psychiatric
- Hypertension
- Cancer
- Renal
- Lung
- Miscellaneous
- Fertility
- Metabolites
- Morphometric
- Blood
- Heart
- All
Return to Forge Tool.
Return to Forge Help.
Autoimmune Diseases
There is a clear enrichment pattern for overlap with hotspots in various myeloid and lymphoid (blood) tissues in the autoimmume diseases. The exact cells involved vary, in was that are compatible with the known differences in aetiologies for the specific diseases.
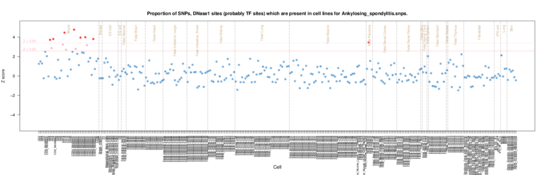
Ankylosing spondylitis
On the Roadmap Epigenome data, there is an enrichment signal for CD14 and CD34 cells. This is consistent with a role for macrophages in the aetiology.
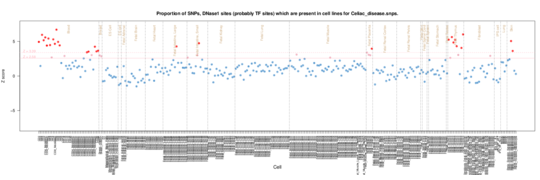
Celiac disease
For Celiac disease, the main enrichment in both datasets is with CD3, CD4 and CD8 cell samples, characteristic of the involvement of thymocuytes and subsequently T cells. The Roadmap Epigenome data also has a strong signal from thymus tissue itself supporting this. This is consistent with the T cell involvement in the disease since the extensive infiltration of CD8(+) T cells in the intestinal mucosa of celiac disease (CD) patients is a hallmark of the disease.
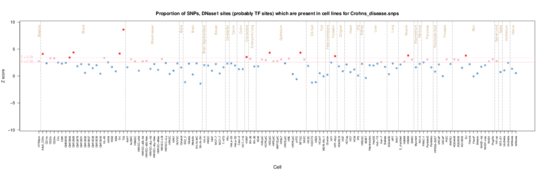
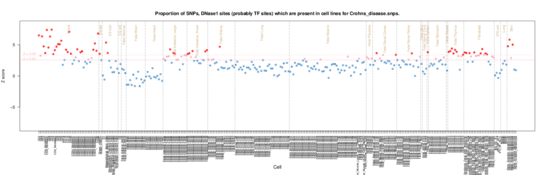
Crohn’s disease
Crohn’s disease GWAS SNPs are enriched for overlap with T helper cells (Th2, CD3,CD4), NK cells (CD56) and some B cell (CD19) and Cytotoxic T cells or NK cells (CD8). The T cell involvement is reflected by a thymus enrichment. Notably there is signal from the small and large intestine and skin/fibroblast cells consistent withteh site of inflammation and altered fibroblast function in the disease,
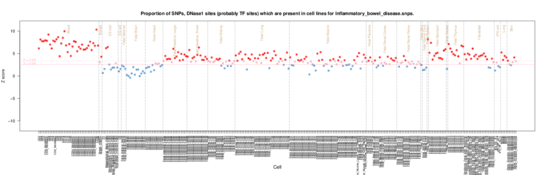
Inflammatory bowel disease
Again as with Crohns there is a T cell and lymphoid signal, but now there is a more general inflammation signal across many tissues reflecting the more gerneral inflammation in inflammatory bowel disease.
Inflammatory bowel disease on Roadmap Epigenome data
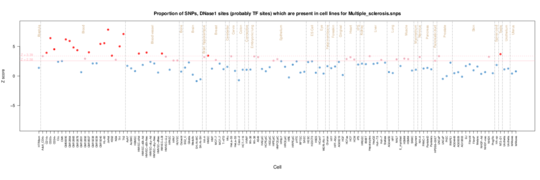
Multiple sclerosis
Again the T cell and thymus inflammatory enrichment is revealed, but now there is enrichment in muscle cells which is intriguing given the effects of neurological damage on muscle function in MS.
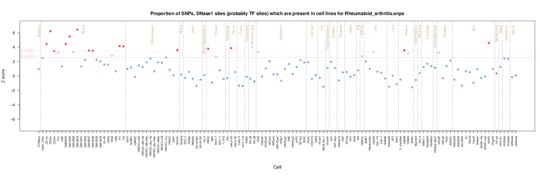
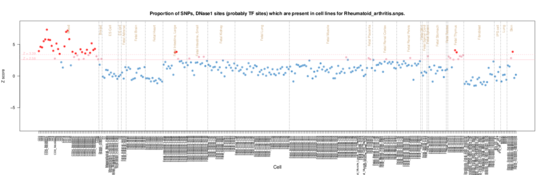
Rheumatoid Arthritis
Rheumatoid Arthritis displays a slightly different pattern of enrichment in the blood cells. Although there is some enrichment in T cells (CD3, CD4 and CD8), thymus cells and CD34+ hematopoietic progenitor cells, the main enrichment is in CD19 B cells. CD19 cell depletion is the target of autoreactive plasma cells in Rheumatoid Arthritis. http://arthritis-research.com/content/14/S5/S1
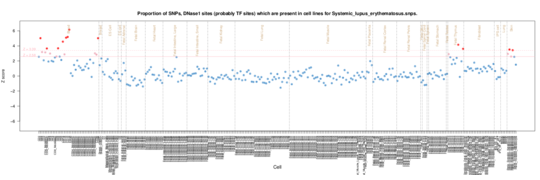
Systemic lupus erythematosus
In SLE an even more pronounced specific enrichment ifn CD19 cells is seen, reflecting the predominently B cell pathology of the disease with it’s anti-nucleic acid antibodies.
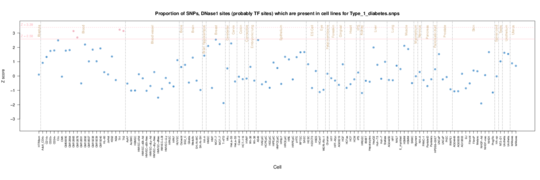
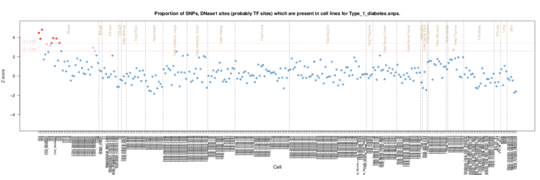
Type 1 diabetes
For Type 1 diabetes the autoimmune signature is not as strong although there is clearly a T cell (CD3, CD4 and CD8) signal in the Roadmap data, consistent with the T cell invasion of the pancreatic iislets in Type 1.
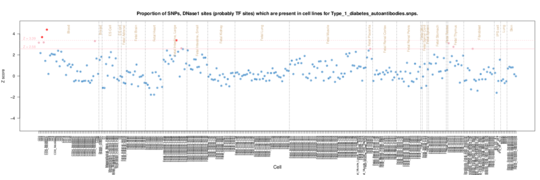
In this autoantibody phenotype in Type 1 diabetes, activated T cell signal can be seen (CD3).
Type 1 diabetes autoantibodies on Roadmap Epigenome data
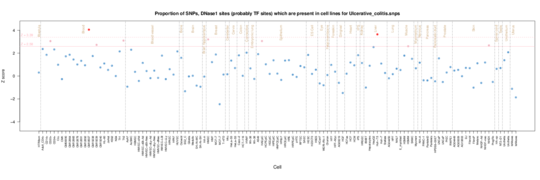
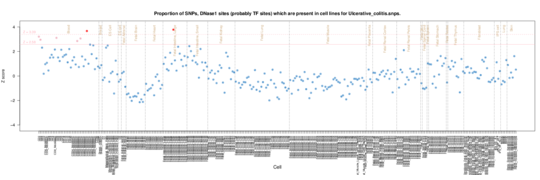
Ulcerative colitis
In Ulcerative Colitis, the other IBD phenotype besides Crohns, the signal is weaker, but is present in CD3, CD34 and CD8 samples.
Psychiatric
Autism spectrum disorder attention deficit-hyperactivity disorder bipolar disorder major depressive disorder and schizophrenia combined
Intriguingly the only signal for this combined psyciatric meta study is in fetal brain samples.
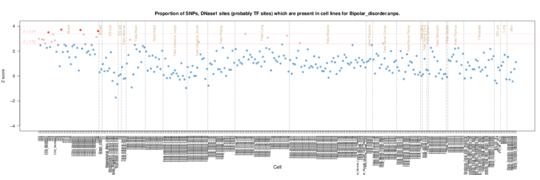
Bipolar disorder
Bipolar disorder SNPs reveal an autoimmune signal. This is puzzling. There have been studies suggesting links between autoimmunity and schizophrenia, but a brief look at the literature suggests it’s not conclusive for bipolar (http://europepmc.org/abstract/MED/20868462).
Hypertension
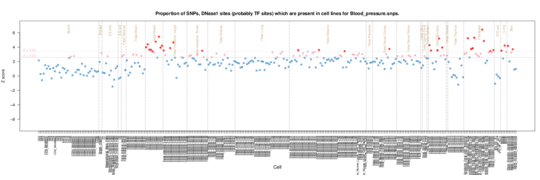
Blood pressure
Enrichment signal in fetal heart, muscle, stomach/intestine and fibroblasts. Maybe makes sense with blood vessle walls being involved.
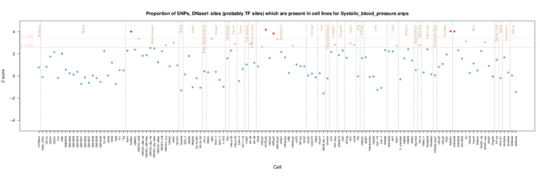
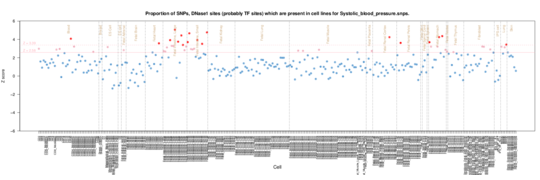
Systolic blood pressure
in Systolic blood pressure the enrichment signal is predominently stomach/intesting inthe Roadmap data with skin and epithelium in ENCODE data.
Cancer
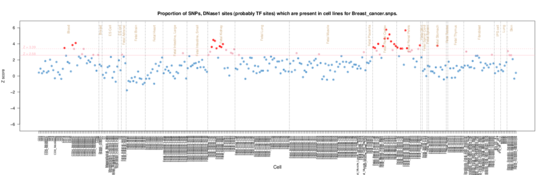
Breast cancer
There is a breast cancer line enrichment in the ENCODE data(MCF-7), but intriguingly a kidney signal in the Roadmap data.
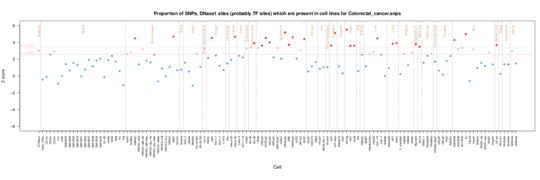
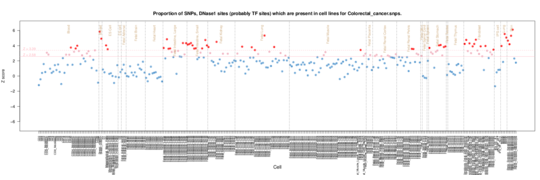
Colorectal cancer
Fetal Stomach/Intesting enrichment in Roadmap data but also epithelium/fibroblasts including breast. In ENCODE cells there is also an epithelium signal but also a more general enrichment perhaps reflecting the prevalence of cancer lines.
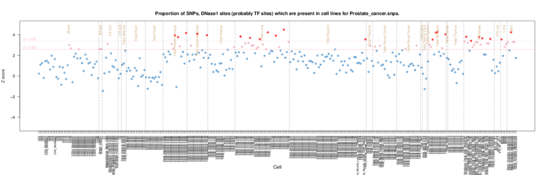
Prostate cancer
Notably enrichment in prostate cancer cells but also Hela (cervical carcinoma) in ENCODE data. Roadmap enrichments highlight fibroblasts and lung/stomach/intesting perhaps reflecting epithelial origins?
Renal
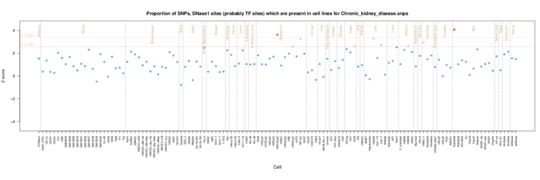
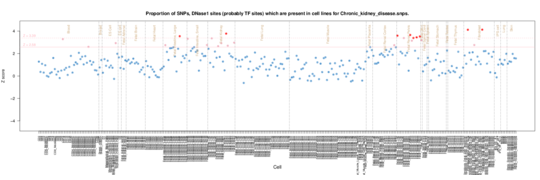
Chronic kidney disease
Enrichment in the renal/kidney samples in the Roadmap data but also fibroblasts.
Lung
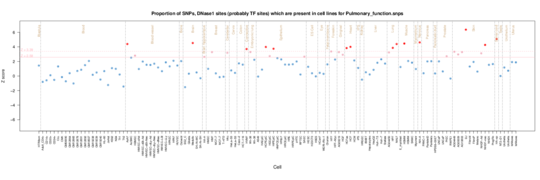
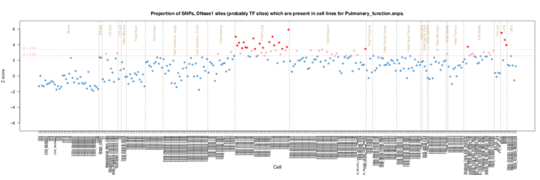
Pulmonary Function
Clear Fetal Lung enrichment in Pulmonary function (spirometry).
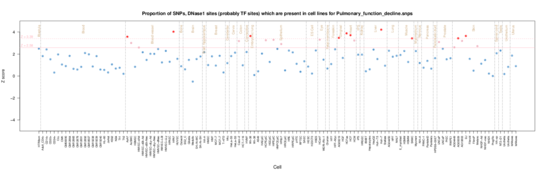
Pulmonary Function Decline
For decline note that muscle is more important than fetal lung pointing towards mechanism.
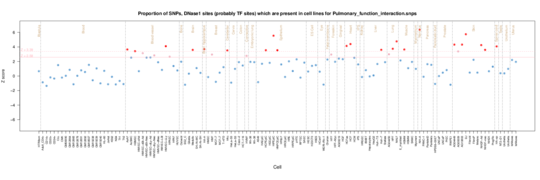
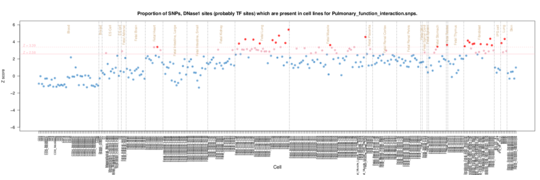
Pulmonary Function Interaction
Not sure on the phenotype here (need to look up the study) but both fetal lung and muscle show enrichment along with fibroblasts.
Miscellaneous
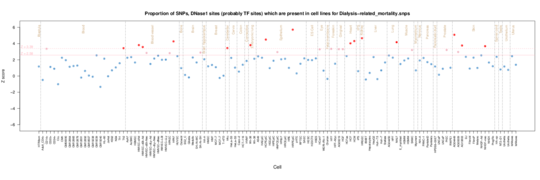
Dialysis-related mortality
Breathing is important when you are on dialysis, as well as fibroblast function.
Fertility
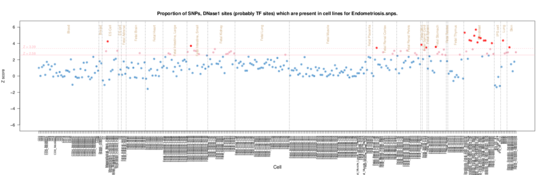
Endometriosis
This is a fibroblast related trait, and this shows in the enrichments,
Metabolites
Urate levels
Mostly Stomach and some Kidney important here.
Fasting glucose-related traits
Fasting glucose-related traits interaction with BMI highlights energy intensive/important tissues, stomach, intestine, muscle. Interesting that pancreas in the ENCODE data is only just enriched.
Morphometric
Height
Almost all tissues are important for height….
Blood
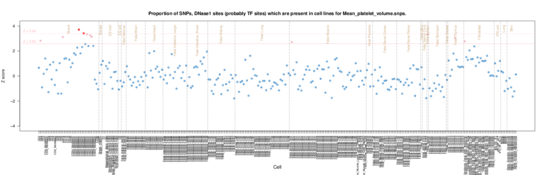
Mean platelet volume
The major signal is from CD34 cells, hematopoietic progenitor cells which makes sense as these ultimately give rise to platelets.
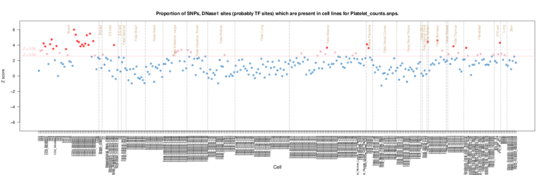
Platelet counts
Again CD34 cells are enriched but there are also alternate B cell, monocytes and T cell blood enrichments and signals from spleen stomach and thymus tissue. The ENCODE data also has some signals from blood vessels, but some odd sporadic enrichments as well.
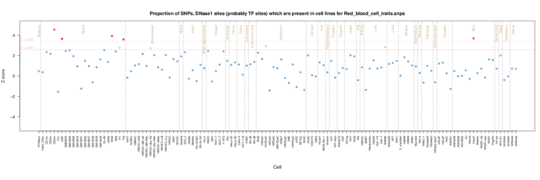
Red blood cell traits
The major signal is from CD34 cells, hematopoietic progenitor cells but spleen and stomach also play a role.
Heart
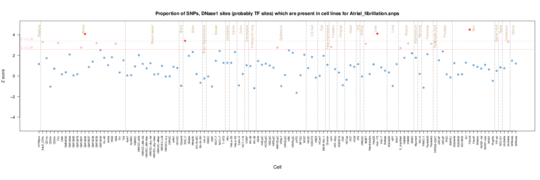
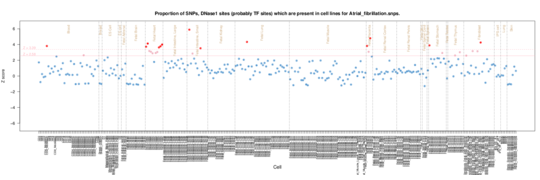
Atrial Fibrillation
Roadmap data shows an enrichment in fetal heart tissue
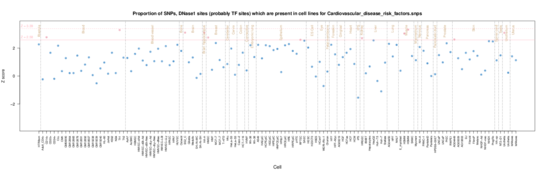
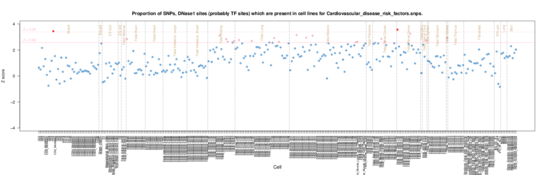
Cardiovascular disease risk factors
These are enriched in renal/kidney tissue, lung and muscle. Nothin really evident in ENCODE data.
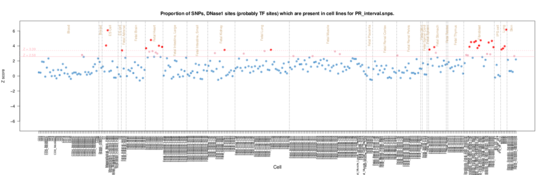
PR interval
ENCODE data picks up a particular enrichment in blood vessels. The Roadmap data picks out fetal heart and fibroblasts.
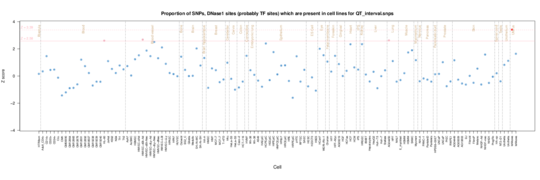
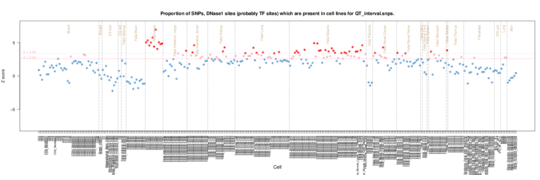
QT interval
The major enrichment is again in fetal heart but in addition we see signal particularly from lung and muscle tissues plus some kidney.
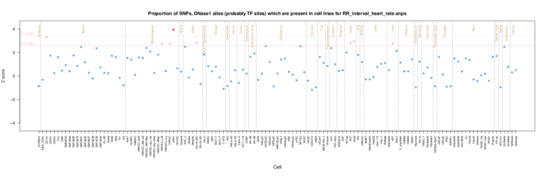
RR interval
For RR rate, the main enrichment is not now in fetal heart but brain, kidney and muscle are also relevant consistent with control of rate being outside the heart itself. Interatingly there is also CD14 and CD19 cell signal (monocyte and B cells).
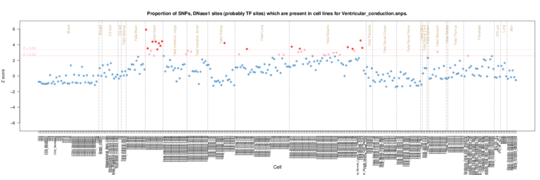
Ventricular conduction on Roadmap Epigenome data
For ventricular conduction the signal is from Fetal Heart and also from Fetal Muscle tissues.
All GWAS SNP hits
Looking across the whole GWAS catalog at once, there is enrichment for almost all tissues. However notably spinal cord and brain shows little enrichment.